My last post focused on the use of the ctree procedure in
the R package party to build classification tree models. These models map each record in a dataset
into one of M mutually exclusive groups, which are characterized by their
average response. For responses coded as
0 or 1, this average may be regarded as an estimate of the probability that a
record in the group exhibits a “positive response.” This interpretation leads to the idea discussed
here, which is to replace this estimate with the size-corrected probability
estimate I discussed in my previous post (Screening for predictive characteristics). Also, as discussed in that post, these
estimates provide the basis for confidence intervals that quantify their
precision, particularly for small groups.
In this post, the basis for these estimates is the R package
PropCIs, which includes several procedures for estimating binomial
probabilities and their confidence intervals, including an implementation of
the method discussed in my previous post.
Specifically, the procedure used here is addz2ci, discussed in Chapter 9
of Exploring Data in Engineering, the Sciences, and Medicine. As
noted in both that discussion and in my previous post, this estimator is
described in a paper by Brown, Cai and DasGupta in 2002, but the documentation
for the PropCIs package cites an earlier paper by Agresti and Coull
(“Approximate is better than exact for interval estimation of binomial
proportions,” in The American Statistician, vol. 52, 1998, pp. 119-126). The essential idea is to modify the classical
estimator, augmenting the counts of 0’s and 1’s in the data by z2/2, where z is
the normal z-score associated with the significance level. As a specific example, z is approximately
1.96 for 95% confidence limits, so this modification adds approximately 2 to
each count. In cases where both of these
counts are large, this correction has negligible effect, so the size-corrected
estimates and their corresponding confidence intervals are essentially
identical with the classical results. In
cases where either the sample is small or one of the possible responses is
rare, these size-corrected results are much more reasonable than the classical
results, which motivated their use both here and in my earlier post.
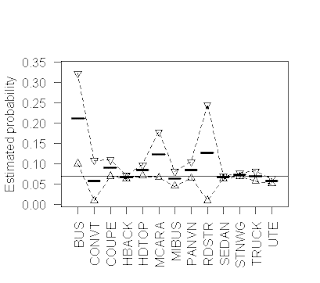
The above plot provides a simple illustration of the results
that can be obtained using the addz2ci procedure, in a case where some groups
are small enough for these size-corrections to matter. More specifically, this plot is based on the
Australian vehicle insurance dataset that I discussed in my last post, and it
characterizes the probability that a policy files a claim (i.e., that the
variable clm has the value 1), for each of the 13 vehicle types included in the
dataset. The heavy horizontal line
segments in this plot represent the size-corrected claim probability estimates
for each vehicle type, while the open triangles connected by dotted lines
represent the upper and lower 95% confidence limits around these probability
estimates, computed as described above.
The solid horizontal line represents the overall claim probability for
the dataset, to serve as a reference value for the individual subset results.
An important observation here is that although this dataset
is reasonably large (there are a total of 67,856 records), the subgroups are
quite heterogeneous in size, spanning the range from 27 records listing “RDSTR”
as the vehicle type to 22,233 listing “SEDAN ”. As a consequence, although the classical and
size-adjusted claim probability estimates and their confidence intervals are
essentially identical for the dataset overall, the extent of this agreement
varies substantially across the different vehicle types. Taking the extremes, the results for the largest
group (“SEDAN”) are, as with the dataset overall, almost identical: the
classical estimate is 0.0665, while the size-adjusted estimate is 0.0664; the
lower 95% confidence limit also differs by one in the fourth decimal place
(classical 0.0631 versus size-corrected 0.0632), and the upper limit is
identical to four decimal places, at 0.0697.
In marked contrast, the classical and size-corrected estimates for the
“RDSTR” group are 0.0741 versus 0.1271, the upper 95% confidence limits are
0.1729 versus 0.2447, and the lower confidence limits are -0.0247 versus
0.0096. Note that in this case, the
lower classical confidence limit violates the requirement that probabilities
must be positive, something that is not possible for the addz2ci confidence
limits (specifically, negative values are less likely to arise, as in this
example, and if they ever do arise, they are replaced with zero, the smallest
feasible value for the lower confidence limit; similarly for upper confidence
limits that exceed 1). As is often the
case, the primary advantage of plotting these results is that it gives us a
much more immediate indication of the relative precision of the probability
estimates, particularly in cases like “RDSTR” where these confidence intervals
are quite wide.
The R code used to generate these results uses both the
addz2ci procedure from the PropCIs package, and the summaryBy procedure from
the doBy package. Specifically, the following
function returns a dataframe with one row for each distinct value of the variable
GroupingVar. The columns of this
dataframe include this value, the total number of records listing this value,
the number of these records for which the binary response variable BinVar is
equal to 1, the lower confidence limit, the upper confidence limit, and the
size-corrected estimate. The function is
called with BinVar, GroupingVar, and the significance level, with a default of
95%. The first two lines of the function
require the doBy and PropCIs packages.
The third line constructs an internal dataframe, passed to the summaryBy
function in the doBy package, which applies the length and sum functions to the
subset of BinVar values defined by each level of GroupingVar, giving the total
number of records and the total number of records with BinVar = 1. The main loop in this program applies the
addz2ci function to these two numbers, for each value of GroupingVar, which
returns a two-element list. The element
$estimate gives the size-corrected probability estimate, and the element
$conf.int is a vector of length 2 with the lower and upper confidence limits
for this estimate. The rest of the
program appends these values to the internal dataframe created by the summaryBy
function, which is returned as the final result. The code listing follows:
BinomialCIbyGroupFunction <- function(BinVar, GroupingVar, SigLevel = 0.95){
#
require(doBy)
require(PropCIs)
#
IntFrame = data.frame(b = BinVar, g = as.factor(GroupingVar))
SumFrame = summaryBy(b ~ g, data = IntFrame, FUN=c(length,sum))
#
n = nrow(SumFrame)
EstVec = vector("numeric",n)
LowVec = vector("numeric",n)
UpVec = vector("numeric",n)
for (i in 1:n){
Rslt = addz2ci(x = SumFrame$b.sum[i],n = SumFrame$b.length[i],conf.level=SigLevel)
EstVec[i] = Rslt$estimate
CI = Rslt$conf.int
LowVec[i] = CI[1]
UpVec[i] = CI[2]
}
SumFrame$LowerCI = LowVec
SumFrame$UpperCI = UpVec
SumFrame$Estimate = EstVec
return(SumFrame)
}
The binary response characterization tools just described can be applied to the results obtained from a classification tree model. Specifically, since a classification tree assigns every record to a unique terminal node, we can characterize the response across these nodes, treating the node numbers as the data groups, analogous to the vehicle body types in the previous example. As a specific illustration, the figure above gives a graphical representation of the ctree model considered in my previous post, built using the ctree command from the party package with the following formula:
Fmla = clm
~ veh_value + veh_body + veh_age + gender + area + agecat
Recall that this formula specifies we want a classification
tree that predicts the binary claim indicator clm from the six variables on the
right-hand side of the tilde, separated by “+” signs. Each of the terminal nodes in the resulting
ctree model is characterized with a rectangular box in the above figure, giving
the number of records in each group (n) and the average positive response (y),
corresponding to the classical claim probability estimate. Note that the product ny corresponds to the
total number of claims in each group, so these products and the group sizes
together provide all of the information we need to compute the size-corrected
claim probability estimates and their confidence limits for each terminal node. Alternatively, we can use the where method associated with the binary
tree object that ctree returns to extract the terminal nodes associated with
each observation. Then, we simply use
the terminal node in place of vehicle body type in exactly the same analysis as
before.
The above figure shows these estimates, in the same format
as the original plot of claim probability broken down by vehicle body type
given earlier. Here, the range of
confidence interval widths is much less extreme than before, but it is still
clearly evident: the largest group (Node 10, with 23,315 records) exhibits the
narrowest confidence interval, while the smallest groups (Node 9, with 1,361
records, and Node 13, with 1,932 records) exhibit the widest confidence
intervals. Despite its small size,
however, the smallest group does exhibit a significantly lower claim
probability than any of the other groups defined by this classification tree
model.
The primary point of this post has been to demonstrate that
binomial confidence intervals can be used to help interpret and explain
classification tree results, especially when displayed graphically as in the
above figure. These displays provide a
useful basis for comparing classification tree models obtained in different
ways (e.g., by different algorithms like rpart and ctree, or by different
tuning parameters for one specific algorithm).
Comparisons of this sort will form the basis for my next post.